Computational Statistics
Lecture 9
Yixuan Qiu
2023-11-08
Optimization
Last Time
We have spent much time on nonsmooth problems
Since they are very common in statistical models
Meanwhile they are considered "hard problems" in convex optimization
So far, the main tool to overcome nonsmoothness is the proximal operator
Seen in proximal gradient descent, Douglas-Rachford splitting, Davis-Yin splitting, etc.
In this lecture we introduce a few other frameworks
Today's Topics
Alternating direction method of multipliers
Coordinate descent
Case studies
Alternating Direction Method of Multipliers
Problem
Another popular framework to deal with nonsmooth or constrained problems is the alternating direction method of multipliers (ADMM). It considers problems of the form:
min
x\in\mathbb{R}^n, z\in\mathbb{R}^m, A\in\mathbb{R}^{p\times n}, B\in\mathbb{R}^{p\times m}, and c\in\mathbb{R}^p.
f(x) and g(z) are convex functions, possibly nonsmooth
Ax+Bz=c represents general linear equality constraints
Examples
Although this problem form is a bit exotic compared with those we have seen in previous lectures
Many familiar problems can be converted into this form by cleverly defining the x and z variables
Lasso:
\min_x\ \frac{1}{2}\Vert b-Ax\Vert^{2}+\lambda\Vert x\Vert_{1}
f(x)=(1/2)\Vert b-Ax\Vert^{2}, g(z)=\lambda\Vert z\Vert_1
Linear constraint x-z=0
Examples
- Least absolute deviations:
\min_x\ \Vert b-Ax \Vert_1
f(x)=0, g(z)=\Vert z\Vert_1
Linear constraint Ax-z=b
Algorithm
ADMM can be expressed as the following algorithm. Given initial values z^{(0)} and u^{(0)}, for k=0,1\ldots, iterate
\begin{align*} x^{(k+1)} & =\underset{x}{\arg\min}\ f(x)+(\rho/2)\Vert Ax+Bz^{(k)}-c+u^{(k)}\Vert^{2}\\ z^{(k+1)} & =\underset{z}{\arg\min}\ g(z)+(\rho/2)\Vert Ax^{(k+1)}+Bz-c+u^{(k)}\Vert^{2}\\ u^{(k+1)} & =u^{(k)}+Ax^{(k+1)}+Bz^{(k+1)}-c \end{align*}
\rho>0 is an arbitray "inverse step size" parameter.
Some Remarks
The x- and z-updates look similar to proximal operators
However, here we have more general quadratic terms
This means that the x- and z-updates are typically more difficult than proximal operators
On the other hand, ADMM does not need to explicitly project to the linear constraint set
These findings largely guide how to define the f and g functions
Relation with DRS
Consider using ADMM to solve the problem
\min_{x}\ f(x)+g(x)\Leftrightarrow\min_{x,z}\ f(x)+g(z)\ \text{ subject to }\ x-z=0
Then the iteration becomes
\begin{align*} x^{(k+1)} & =\mathbf{prox}_{(1/\rho)f}(z^{(k)}-u^{(k)})\\ z^{(k+1)} & =\mathbf{prox}_{(1/\rho)g}(x^{(k+1)}+u^{(k)})\\ u^{(k+1)} & =u^{(k)}+x^{(k+1)}-z^{(k+1)} \end{align*}
This is equivalent to DRS, by slightly reparameterizing the variables.
Implementation Details
Many implementation details can be found in the main reference [1]
Stopping criterion
Selection of the \rho parameter
Closed-form expressions for specific problems
Convergence Property [2]
The sequence (x^{(k)},z^{(k)},u^{(k)}) satisfy
\small\Vert A(x^{(k+1)}-x^{(k)})\Vert^2+\Vert B(z^{(k+1)}-z^{(k)})\Vert^2+\Vert u^{(k+1)}-u^{(k)}\Vert^2=o(1/k).
This is one of the most general convergence results on ADMM.
Many other faster rates exist by imposing stronger conditions on f(x) and g(z), e.g. smoothness, strong convexity, etc.
Coordinate Descent
Motivation
One more idea to overcome nonsmoothness is to directly find the optimal point of nonsmooth problems
Of course, this is unrealistic for general problems
But in many cases this is possible for univariate functions
Example
Lasso model \min_\beta\ \ell(\beta):=\min_\beta\ \frac{1}{2n}\Vert y-X\beta\Vert^{2}+\lambda\Vert \beta\Vert_{1}
Solving the whole \beta vector is hard
However, assume we fix all elements of \beta except \beta_i
Then we can actually derive the optimal \beta_i in closed form
Example
- Recall that in Lecture 7 we have shown
f(x^*)=\min_{x}\,f(x)\Leftrightarrow 0\in \partial f(x^*)
- View \tilde{\ell}(\beta_i)\equiv\ell(\beta) as a function of \beta_i, then
\partial\tilde{\ell}(\beta_{i})=n^{-1}\Vert X_{i}\Vert^2\beta_{i}+n^{-1}X_{i}'(X_{-i}\beta_{-i}-y)+\lambda\partial|\beta_{i}|,
X_i is the i-column of X
X_{-i} is the matrix formed by removing X_i from X
\beta_{-i} is the vector formed by removing \beta_i from \beta
Example
- 0\in \partial\tilde{\ell}(\beta_{i}) gives the solution
\beta_{i}=\mathcal{S}_{n\lambda/\Vert X_{i}\Vert^{2}}\left(\frac{X_{i}'(y-X_{-i}\beta_{-i})}{\Vert X_{i}\Vert^{2}}\right)
Coordinate Descent
These findings motivate the coordinate descent (CD) algorithm. Consider the problem
\min_x\ f(x):=\min_x\ f(x_1,\ldots,x_n) One common type of CD algorithm proceeds as follows. Given initial value x^{(0)}, for k=0,1,\ldots, iterate:
x_{i}^{(k+1)}=\underset{x_{i}}{\arg\min}\ f(x_{1}^{(k+1)},\ldots,x_{i-1}^{(k+1)},x_{i},x_{i+1}^{(k)},\ldots,x_{n}^{(k)}) for i=1,\ldots,n within each k-outer-iteration.
This is typically called the cyclic CD algorithm.
Variants
Another popular variant is to randomly select a coordinate to update. Given initial value x^{(0)}, for k=0,1,\ldots, do:
Randomly select i_k from \{1,2,\ldots,n\} with equal probabilities
Fixing other coordinates and update
\begin{align*} x_{i_{k}}^{(k+1)} & =\underset{x_{i_{k}}}{\arg\min}\ f(x_{1}^{(k)},\ldots,x_{i_{k}-1}^{(k)},x_{i_{k}},x_{i_{k}+1}^{(k)},\ldots,x_{n}^{(k)})\\ x_{j}^{(k+1)} & =x_{j}^{(k)},\quad j\neq i_{k} \end{align*}
In the literature, this method is sometimes called randomized CD.
Variants
Many other variants exist:
Permutation CD: for each k, use a random permutation of \{1,2,\ldots,n\} to define the order of coordinate updates
Block CD: each time update a block of coordinates simultaneously, instead of a single coordinate
Coordinate proximal gradient descent: use proximal operators to update coordinates
...
Convergence Property [3]
Unfortunately, CD does not always converge to an optimal point for nonsmooth f. Instead, consider the problem
f(x)=g(x)+\sum_{i=1}^n h_i(x_i),
where g(x) is convex and differentiable, and each h_i(x_i) is convex, possibly nonsmooth.
[3] shows that with very mild regularity conditions, the CD iterates x^{(k)} converge to an optimal point of f(x).
The result holds for block CD and arbitrary order of cycle through coordinates.
Convergence Property [4]
There exist many convergence results for different variants of CD based on different assumptions on f(x). Below is one representative result with weak assumptions.
Assume that g(x) is coordinate-wise L_i-smooth, and define \small R_{0}=\max_{y}\max_{x^{\\*}\in\mathcal{X}^{\\*}}\left\{ \left(\sum_{i=1}^{n}L_{i}(y_{i}-x_{i}^{\\*})^{2}\right)^{1/2}:f(y)\le f(x^{(0)})\right\}, where \mathcal{X}^* denotes the optimal set.
(See next slide)
Convergence Property [4]
Consider the randomized CD algorithm. Fix a confidence parameter 0<\rho<1 and an accuracy level \varepsilon < \min\{R_0^2,f(x^{(0)})-f(x^*)\}. Then with
k\ge \frac{2nR_{0}^{2}}{\varepsilon}\log\frac{f(x^{(0)})-f(x^{*})}{\varepsilon\rho}, we have f(x^{(k)})-f(x^*)\le \varepsilon with probability at least 1-\rho.
Convergence with Strong Convexity [4]
For simplicity suppose L_i\equiv L, and assume that g(x) is also strongly convex with parameter m>0. Fix a confidence parameter 0<\rho<1 and an accuracy level \varepsilon>0. If
k\ge\frac{4Ln}{m}\log\frac{f(x^{(0)})-f(x^{*})}{\varepsilon\rho},
then f(x^{(k)})-f(x^*)\le \varepsilon with probability at least 1-\rho.
Case Study
Lasso
We use the lasso model \min_\beta\ \ell(\beta):=\min_\beta\ \frac{1}{2n}\Vert y-X\beta\Vert^{2}+\lambda\Vert \beta\Vert_{1} to study the convergence property and computational efficiency of several algorithms we have covered in previous lectures.
Derive the update rules for several algorithms
Study the change of loss function versus iteration number
Study the change of loss function versus running time
Simulate Data Set
The design matrix is a simulated sparse matrix, but we tentatively use its dense version.
library(Matrix)set.seed(123)n = 500p = 1000s = 30xsp = rsparsematrix(n, p, density = 0.1)x = as.matrix(xsp)beta = c(rnorm(s), numeric(p - s))y = as.numeric(xsp %*% beta) + 0.1 * rnorm(n)lam = 0.001objfn = function(beta, x, y, lam){ 0.5 * mean((y - as.numeric(x %*% beta))^2) + lam * sum(abs(beta))}Compute Optimal Value
Use the celebrated glmnet package to compute the optimal value.
library(glmnet)res = glmnet(x, y, lambda = lam, thresh = 1e-15, standardize = FALSE, intercept = FALSE)beta_opt = as.numeric(res$beta)head(beta_opt)## [1] -0.3862279 1.4304118 -0.2206914 1.1256892 0.1704705 0.8657187tail(beta_opt)## [1] 0.000000000 -0.003938278 0.000000000 0.000000000 0.000000000## [6] 0.008219107obj_opt = objfn(beta_opt, x, y, lam)obj_opt## [1] 0.02782515Optimization
A general interface for running optimization algorithms.
run_opt = function(x, y, lam, init_state, update_rule, maxit = 1001, ...){ ...}init_state()andupdate_rule()are two function arguments to characterize the optimization algorithminit_state(x, y, lam, ...)is used to create initial state of the algorithmupdate_rule(beta, iter, x, y, lam, state)returns the updated \beta and state,list(beta, state)
Optimization
A general interface for running optimization algorithms.
run_opt = function(x, y, lam, init_state, update_rule, maxit = 1001, ...){ p = ncol(x) beta_hat = numeric(p) losses = c() # Initialization state = init_state(x, y, lam, ...) # Main loop for(i in 1:maxit) { res = update_rule(beta_hat, i - 1, x, y, lam, state) beta_hat = res$beta state = res$state loss = objfn(beta_hat, x, y, lam) losses = c(losses, loss) if((i - 1) %% 100 == 0) cat(sprintf("Iter %d, loss = %.6f\n", i - 1, loss)) } list(beta_hat = beta_hat, losses = losses)}Subgradient Method
Choose \tilde{\nabla}\ell(\beta)=(1/n)X'(X\beta-y)+\lambda\cdot\mathrm{sign}(\beta)
Update rule: \beta^{(k+1)}=\beta^{(k)}-\alpha_{k}\tilde{\nabla}\ell(\beta^{(k)})
Step size \alpha_k=\alpha_0/\sqrt{k+1}
Implementation
init_subgrad = function(x, y, lam, alpha0 = 1){ list(alpha0 = alpha0)}update_subgrad = function(beta, k, x, y, lam, state){ n = nrow(x) alpha0 = state$alpha0 alphak = alpha0 / sqrt(k + 1) Xtr = crossprod(x, as.numeric(x %*% beta) - y) subgrad = as.numeric(Xtr) / n + lam * sign(beta) new_beta = beta - alphak * subgrad list(beta = new_beta, state = state)}Result
res_subgrad1 = run_opt( x, y, lam, init_subgrad, update_subgrad, alpha0 = 1)## Iter 0, loss = 0.770908## Iter 100, loss = 0.084973## Iter 200, loss = 0.074552## Iter 300, loss = 0.070417## Iter 400, loss = 0.067906## Iter 500, loss = 0.066074## Iter 600, loss = 0.064626## Iter 700, loss = 0.063428## Iter 800, loss = 0.062394## Iter 900, loss = 0.061477## Iter 1000, loss = 0.060661head(res_subgrad1$beta)## [1] -2.174641e-01 9.316010e-01 -1.026982e-01 6.878443e-01 1.129936e-05## [6] 2.784390e-01tail(res_subgrad1$beta)## [1] -3.352589e-02 -5.879592e-02 -1.432275e-06 -3.051985e-02 5.442229e-02## [6] 1.141592e-01Result
res_subgrad10 = run_opt( x, y, lam, init_subgrad, update_subgrad, alpha0 = 10)## Iter 0, loss = 8.881744## Iter 100, loss = 0.049232## Iter 200, loss = 0.040807## Iter 300, loss = 0.036166## Iter 400, loss = 0.032993## Iter 500, loss = 0.030828## Iter 600, loss = 0.029429## Iter 700, loss = 0.028636## Iter 800, loss = 0.028227## Iter 900, loss = 0.028043## Iter 1000, loss = 0.027961head(res_subgrad10$beta)## [1] -0.3913635 1.4284850 -0.2179803 1.1101840 0.1683580 0.8471952tail(res_subgrad10$beta)## [1] -2.979032e-04 -3.842055e-03 2.795830e-04 -9.758347e-05 -1.092878e-04## [6] 1.166201e-02Proximal Gradient Descent
Update rule: \beta^{(k+1)}=\mathcal{S}_{\alpha\lambda}\left(\beta^{(k)}-(\alpha/n)X'(X\beta^{(k)}-y)\right)
Step size \alpha=1/\lambda_\max(n^{-1}X'X)=n/\lambda_\max(X'X)
Implementation
library(RSpectra)# Soft-thresholding functionsoft_thresh = function(x, a){ sign(x) * pmax(0, abs(x) - a)}init_ista = function(x, y, lam){ # Compute the largest eigenvalue of X'X alpha = nrow(x) / svds(x, k = 1, nu = 0, nv = 0)$d^2 list(alpha = alpha)}update_ista = function(beta, k, x, y, lam, state){ n = nrow(x) alpha = state$alpha Xtr = crossprod(x, as.numeric(x %*% beta) - y) grad = as.numeric(Xtr) / n new_beta = soft_thresh(beta - alpha * grad, alpha * lam) list(beta = new_beta, state = state)}Result
res_ista = run_opt(x, y, lam, init_ista, update_ista)## Iter 0, loss = 0.494847## Iter 100, loss = 0.044493## Iter 200, loss = 0.032321## Iter 300, loss = 0.028040## Iter 400, loss = 0.027830## Iter 500, loss = 0.027825## Iter 600, loss = 0.027825## Iter 700, loss = 0.027825## Iter 800, loss = 0.027825## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_ista$beta)## [1] -0.3862280 1.4304114 -0.2206905 1.1256887 0.1704708 0.8657158tail(res_ista$beta)## [1] 0.000000000 -0.003938134 0.000000000 0.000000000 0.000000000## [6] 0.008219716Accelerated Proximal Gradient Descent
- Update rule:
\begin{align*} u^{(k+1)} & =\beta^{(k)}+\frac{k-1}{k+2}(\beta^{(k)}-\beta^{(k-1)})\\ \beta^{(k+1)} & =\mathcal{S}_{\alpha\lambda}\left(u^{(k)}-(\alpha/n)X'(Xu^{(k)}-y)\right) \end{align*}
- Step size \alpha=1/\lambda_\max(n^{-1}X'X)=n/\lambda_\max(X'X)
Implementation
init_fista = function(x, y, lam){ p = ncol(x) alpha = nrow(x) / svds(x, k = 1, nu = 0, nv = 0)$d^2 list(alpha = alpha, old_beta = numeric(p), betau = numeric(p))}update_fista = function(beta, k, x, y, lam, state){ n = nrow(x) alpha = state$alpha state$betau = if(k <= 1) beta else beta + (k - 1) / (k + 2) * (beta - state$old_beta) Xtr = crossprod(x, as.numeric(x %*% state$betau) - y) grad = as.numeric(Xtr) / n new_beta = soft_thresh(state$betau - alpha * grad, alpha * lam) state$old_beta = beta list(beta = new_beta, state = state)}Result
res_fista = run_opt(x, y, lam, init_fista, update_fista)## Iter 0, loss = 0.494847## Iter 100, loss = 0.027830## Iter 200, loss = 0.027825## Iter 300, loss = 0.027825## Iter 400, loss = 0.027825## Iter 500, loss = 0.027825## Iter 600, loss = 0.027825## Iter 700, loss = 0.027825## Iter 800, loss = 0.027825## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_fista$beta)## [1] -0.3862278 1.4304116 -0.2206913 1.1256895 0.1704706 0.8657180tail(res_fista$beta)## [1] 0.000000000 -0.003938369 0.000000000 0.000000000 0.000000000## [6] 0.008219289Douglas-Rachford Splitting
- Let g(\beta)=\lambda\Vert \beta\Vert_{1}, f(\beta)=\frac{1}{2n}\Vert y-X\beta\Vert^{2}. Then
\begin{align*} \mathbf{prox}_{\alpha g}(z) & =\mathcal{S}_{\alpha\lambda}(z)\\ \mathbf{prox}_{\alpha h}(\beta) & =(I+(\alpha/n)X'X)^{-1}(\beta+(\alpha/n)X'y) \end{align*}
- Update rule:
\begin{align*} \beta^{(k+1)} & =\mathcal{S}_{\alpha\lambda}\left(u^{(k)}\right)\\ u^{(k+1)} & =u^{(k)}+A^{-1}(2\beta^{(k+1)}-u^{(k)}+(\alpha/n)X'y)-\beta^{(k+1)} \end{align*} where A=I+(\alpha/n)X'X
Implementation
chol_solve = function(R, b){ y = backsolve(R, b, transpose = TRUE) # R'*y = b x = backsolve(R, y) # R*x = y x}init_drs = function(x, y, lam, alpha = 1){ n = nrow(x) p = ncol(x) scaled_xty = alpha / n * as.numeric(crossprod(x, y)) R = chol(diag(p) + alpha / n * crossprod(x)) list(alpha = alpha, sxty = scaled_xty, R = as.matrix(R), betau = numeric(p))}update_drs = function(beta, k, x, y, lam, state){ n = nrow(x) alpha = state$alpha new_beta = soft_thresh(state$betau, alpha * lam) rhs = 2 * new_beta - state$betau + state$sxty state$betau = state$betau + chol_solve(state$R, rhs) - new_beta list(beta = new_beta, state = state)}Result
res_drs1 = run_opt(x, y, lam, init_drs, update_drs, alpha = 1)## Iter 0, loss = 1.454710## Iter 100, loss = 0.053128## Iter 200, loss = 0.041675## Iter 300, loss = 0.034461## Iter 400, loss = 0.029864## Iter 500, loss = 0.028097## Iter 600, loss = 0.027848## Iter 700, loss = 0.027828## Iter 800, loss = 0.027826## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_drs1$beta)## [1] -0.3863738 1.4304559 -0.2205224 1.1254740 0.1704870 0.8652928tail(res_drs1$beta)## [1] 0.000000000 -0.003902763 0.000000000 0.000000000 0.000000000## [6] 0.008261868Result
res_drs10 = run_opt(x, y, lam, init_drs, update_drs, alpha = 10)## Iter 0, loss = 1.454710## Iter 100, loss = 0.027825## Iter 200, loss = 0.027825## Iter 300, loss = 0.027825## Iter 400, loss = 0.027825## Iter 500, loss = 0.027825## Iter 600, loss = 0.027825## Iter 700, loss = 0.027825## Iter 800, loss = 0.027825## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_drs10$beta)## [1] -0.3862278 1.4304117 -0.2206913 1.1256895 0.1704706 0.8657182tail(res_drs10$beta)## [1] 0.000000000 -0.003938377 0.000000000 0.000000000 0.000000000## [6] 0.008219268ADMM
Let f(\beta)=\lambda\Vert \beta\Vert_{1}, g(\beta)=\frac{1}{2n}\Vert y-X\beta\Vert^{2}. Then
Update rule:
\begin{align*} \beta^{(k+1)} & =\mathcal{S}_{\lambda/\rho}\left(z^{(k)}-u^{(k)}\right)\\ z^{(k+1)} & =A^{-1}(\beta^{(k+1)}+u^{(k)}+(n\rho)^{-1}X'y)\\ u^{(k+1)} & =u^{(k)}+\beta^{(k+1)}-z^{(k+1)} \end{align*} where A=I+(n\rho)^{-1}X'X
- A reparameterized DRS
Implementation
init_admm = function(x, y, lam, rho = 1){ n = nrow(x) p = ncol(x) scaled_xty = as.numeric(crossprod(x, y)) / n / rho R = chol(diag(p) + crossprod(x) / n / rho) list(rho = rho, sxty = scaled_xty, R = as.matrix(R), betaz = numeric(p), betau = numeric(p))}update_admm = function(beta, k, x, y, lam, state){ n = nrow(x) rho = state$rho new_beta = soft_thresh(state$betaz - state$betau, lam / rho) rhs = new_beta + state$betau + state$sxty state$betaz = chol_solve(state$R, rhs) state$betau = state$betau + new_beta - state$betaz list(beta = new_beta, state = state)}Result
res_admm1 = run_opt(x, y, lam, init_admm, update_admm, rho = 1)## Iter 0, loss = 1.454710## Iter 100, loss = 0.053098## Iter 200, loss = 0.041664## Iter 300, loss = 0.034453## Iter 400, loss = 0.029859## Iter 500, loss = 0.028096## Iter 600, loss = 0.027848## Iter 700, loss = 0.027828## Iter 800, loss = 0.027826## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_admm1$beta)## [1] -0.3863737 1.4304559 -0.2205226 1.1254743 0.1704871 0.8652934tail(res_admm1$beta)## [1] 0.000000000 -0.003902788 0.000000000 0.000000000 0.000000000## [6] 0.008261821Result
res_admm0.1 = run_opt(x, y, lam, init_admm, update_admm, rho = 0.1)## Iter 0, loss = 1.454710## Iter 100, loss = 0.027825## Iter 200, loss = 0.027825## Iter 300, loss = 0.027825## Iter 400, loss = 0.027825## Iter 500, loss = 0.027825## Iter 600, loss = 0.027825## Iter 700, loss = 0.027825## Iter 800, loss = 0.027825## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_admm0.1$beta)## [1] -0.3862278 1.4304117 -0.2206913 1.1256895 0.1704706 0.8657182tail(res_admm0.1$beta)## [1] 0.000000000 -0.003938377 0.000000000 0.000000000 0.000000000## [6] 0.008219268Coordinate Descent
Each iteration contains a full cycle of \beta_i updates
Update rule:
\beta_{i}\leftarrow\mathcal{S}_{n\lambda/\Vert X_{i}\Vert^{2}}\left(\frac{X_{i}'(y-X_{-i}\beta_{-i})}{\Vert X_{i}\Vert^{2}}\right)
- A "cleverer" scheme is to note that
\beta_{i}\leftarrow\mathcal{S}_{n\lambda/\Vert X_{i}\Vert^{2}}\left(\beta_i+\frac{X_{i}'r}{\Vert X_{i}\Vert^{2}}\right),\quad r=y-X\beta
- Then update r\leftarrow r - (\beta_i^{new} - \beta_i^{old})X_i
Implementation
# Soft-thresholding functionsoft_thresh = function(x, a){ sign(x) * pmax(0, abs(x) - a)}init_cd = function(x, y, lam){ xi_square = colSums(x^2) thresh = nrow(x) * lam / xi_square list(xi_square = xi_square, thresh = thresh)}update_cd = function(beta, k, x, y, lam, state){ new_beta = beta # betai <- S(betai + xi' * r / ||xi||^2, t), r = y - x * beta r = y - c(x %*% beta) for(i in seq_along(new_beta)) { old = new_beta[i] newb = old + sum(x[, i] * r) / state$xi_square[i] thresh = state$thresh[i] new_beta[i] = soft_thresh(newb, thresh) r = r - sum(new_beta[i] - old) * x[, i] } list(beta = new_beta, state = state)}Result
res_cd = run_opt(x, y, lam, init_cd, update_cd)## Iter 0, loss = 0.046708## Iter 100, loss = 0.027825## Iter 200, loss = 0.027825## Iter 300, loss = 0.027825## Iter 400, loss = 0.027825## Iter 500, loss = 0.027825## Iter 600, loss = 0.027825## Iter 700, loss = 0.027825## Iter 800, loss = 0.027825## Iter 900, loss = 0.027825## Iter 1000, loss = 0.027825head(res_cd$beta)## [1] -0.3862278 1.4304117 -0.2206913 1.1256895 0.1704706 0.8657182tail(res_cd$beta)## [1] 0.000000000 -0.003938377 0.000000000 0.000000000 0.000000000## [6] 0.008219268Comparison
K = 1000gdat = data.frame( iter = rep(1:K, times = 8), loss = c(res_subgrad1$losses[1:K], res_subgrad10$losses[1:K], res_ista$losses[1:K], res_fista$losses[1:K], res_drs1$losses[1:K], res_drs10$losses[1:K], res_admm1$losses[1:K], res_cd$losses[1:K]), method = rep(c("Subgrad α=1", "Subgrad α=10", "ISTA", "FISTA", "DRS α=1", "DRS α=10", "ADMM ρ=1", "CD"), each = K))Comparison
library(ggplot2)ggplot(gdat, aes(x = iter, y = log10(loss - obj_opt))) + geom_line(aes(color = method)) + scale_color_hue("Method") + xlab("Iteration") + ylab("Log-error") + theme_bw()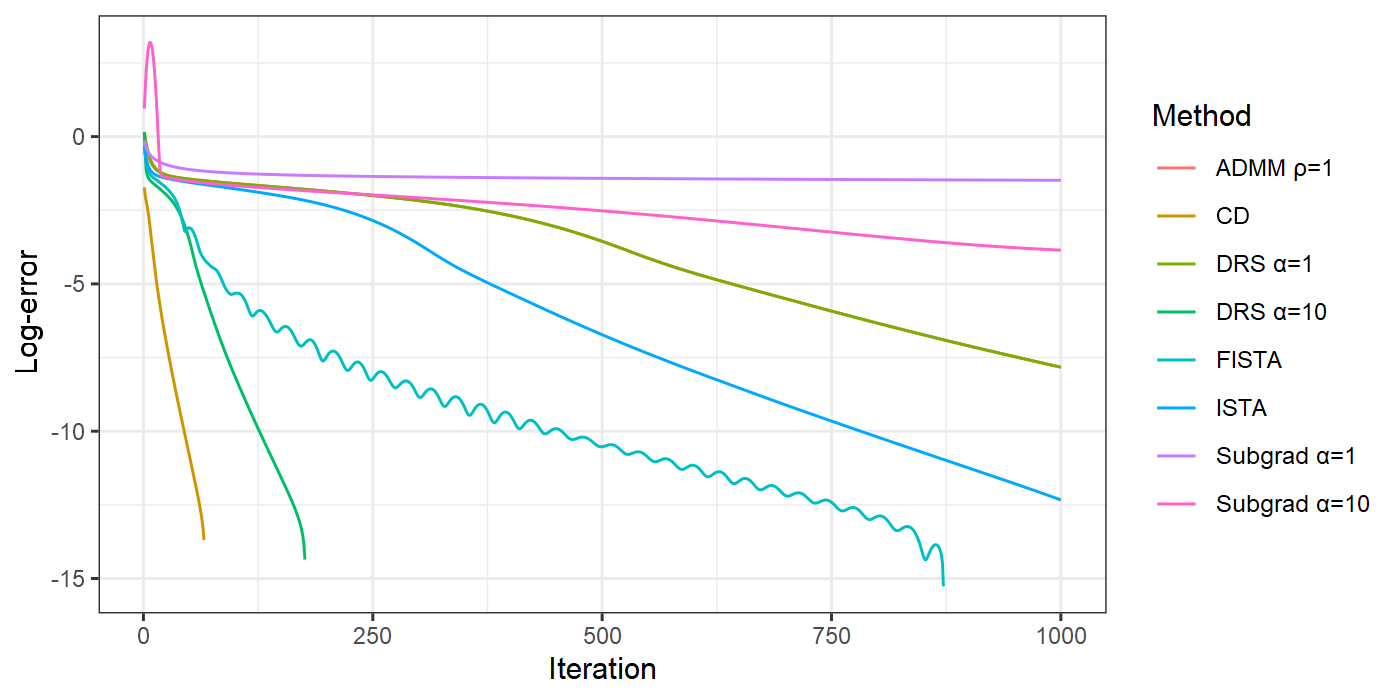
More Considerations
The previous comparison is neither rigorous nor complete, and more works need to be done:
The plot only shows error v.s. iteration, but not error v.s. computing time
The direct factorization of A=I+(\alpha/n)X'X is not optimal (recall that p>n)
Design of algorithms when X is sparse (i.e., operating on the
xspobject)
Left as after-class exercises.
References
[1] Stephen Boyd et al. (2011). Distributed optimization and statistical learning via the alternating direction method of multipliers. Foundations and Trends® in Machine learning.
[2] Deng Wei et al. (2017). Parallel multi-block ADMM with O(1/k) convergence. Journal of Scientific Computing.
[3] Paul Tseng (2001). Convergence of a block coordinate descent method for nondifferentiable minimization. Journal of optimization theory and applications.
[4] Peter Richtárik and Martin Takáč (2014). Iteration complexity of randomized block-coordinate descent methods for minimizing a composite function. Mathematical Programming.