Computational Statistics
Lecture 4
Yixuan Qiu
2023-09-27
Numerical Linear Algebra
Today's Topics
Conjugate gradient method
Eigenvalue computation
Conjugate Gradient Method
Direct Methods
All the methods introduced so far can be categorized as direct methods to solve linear systems
Meaning that the exact solution to Ax=b can be computed within a finite number of operations (in exact arithmetic)
Cons: too expensive to handle very large linear systems
Iterative Methods
Computes a sequence of approximate solutions x(k) that converges to the exact solution
Stops when the precision is sufficient
Do not request more precision than is needed
In each iteration, the computation is typically cheap, e.g., matrix-vector multiplication v→Av
Especially efficient for sparse matrices
Conjugate Gradient Method
Conjugate gradient method (CG) is a very special linear system solver
It is a direct method used as an iterative one
Aims to solve the linear system Ax=b when A is positive definite (p.d.)
Conjugate Gradient Method
Only uses the matrix-vector multiplication v→Av
Obtains the exact solution after n steps
Converges fast under some conditions
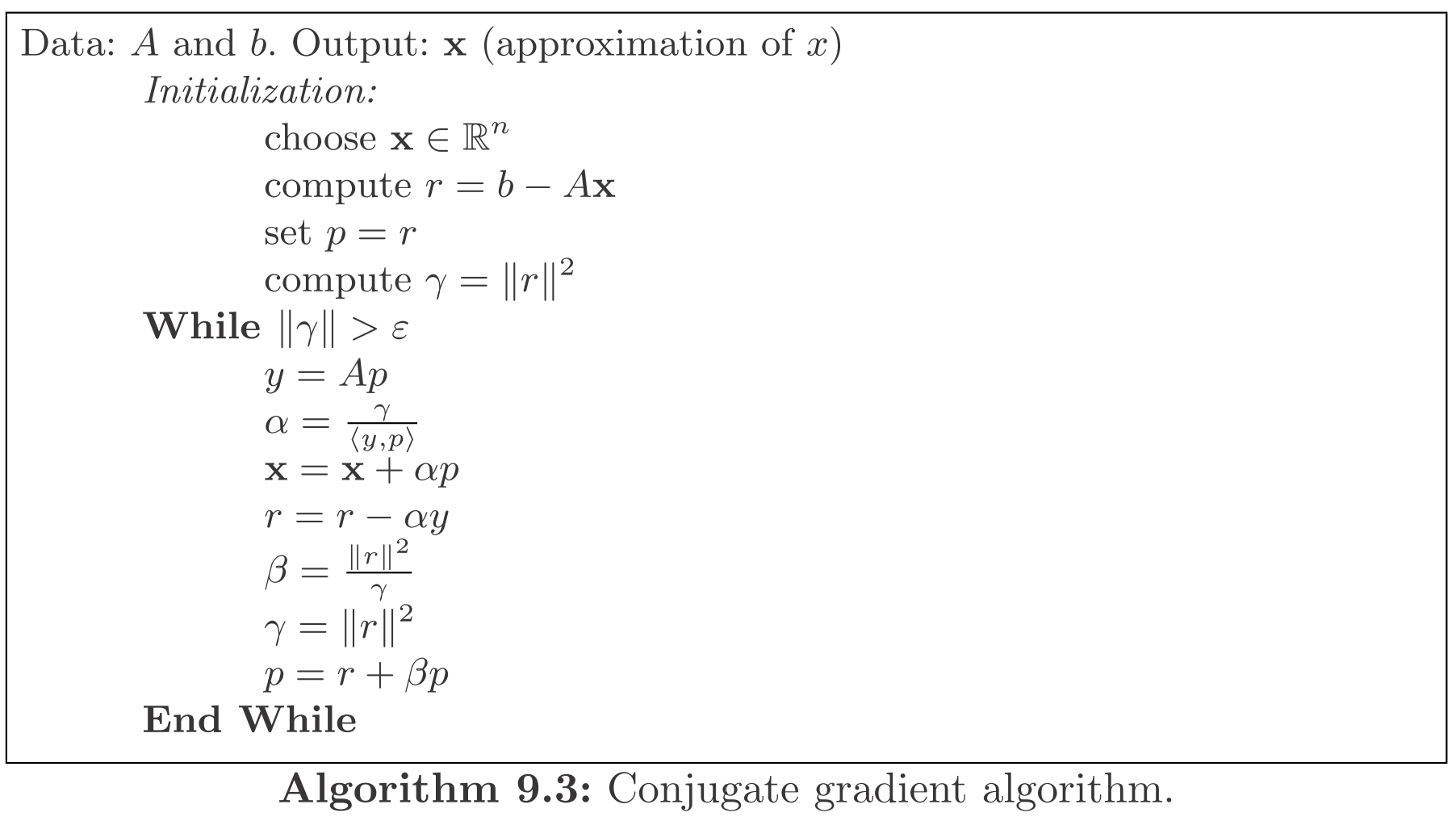
Algorithm [2]
Convergence [2]
In exact arithmetic, the algorithm converges to the solution of the linear system Ax=b in at most n iterations.
Convergence [2]
Let A be a p.d. matrix and denote by x∗ the solution to Ax=b. Let x(k) be the sequence of approximate solutions produced by the conjugate gradient method. Then
‖x(k)−x∗‖2≤2√κ(√κ−1√κ+1)k‖x0−x∗‖2, where κ is the condition number of A (defined later).
Condition Number
For any matrix A, the condition number κ(A) is defined as κ(A)=μmax, where \mu_{\max} and \mu_{\min} are the largest and smallest singular values, respectively
For a p.d. matrix A, \kappa(A)=\lambda_{\max}(A)/\lambda_{\min}(A), where \lambda_{\max} and \lambda_{\min} are the largest and smallest eigenvalues, respectively
\kappa(A)\ge 1
For any orthogonal matrix Q, \kappa(Q)=1 and \kappa(QA)=\kappa(AQ)=\kappa(A)
Insights
- We say a matrix A is well-conditioned if \kappa(A) is small (i.e., \kappa(A)\approx 1)
- Otherwise we say A is ill-conditioned (i.e., \kappa(A)\gg 1)
If a p.d. matrix A has almost equal eigenvalues, then \kappa(A)\approx 1 and \frac{\sqrt{\kappa}-1}{\sqrt{\kappa}+1}\approx 0. CG will have extremely fast convergence
If A almost loses positive definiteness, \lambda_{\min}(A)\approx 0, then \kappa(A)\gg 1, and CG almost has no progress
Implementation
- A direct "translation" of the algorithm gives
cg = function(A, b, x0 = rep(0, length(b)), eps = 1e-6){ m = length(b) x = x0 p = r = b - A %*% x r2 = sum(r^2) errs = c() for(i in 1:m) { Ap = A %*% p alpha = r2 / sum(p * Ap) x = x + alpha * p r = r - alpha * Ap r2_new = sum(r^2) err = sqrt(r2_new) errs = c(errs, err) if(err < eps) break beta = r2_new / r2 p = r + beta * p r2 = r2_new } list(x = x, errs = errs, niter = i)}Implementation
We test on a simulated matrix
However, the algorithm does not seem to converge, and the error is quite large
set.seed(123)n = 100M = matrix(0.1 * rnorm(n^2), n)A = crossprod(M)b = rnorm(n)sol = cg(A, b, eps = 1e-12)sol$niter## [1] 100max(abs(A %*% sol$x - b))## [1] 0.5763024Implementation
- We can find that A is ill-conditioned
kappa(A)## [1] 1164091- Adding a positive number to the diagonal results in much smaller condition number
kappa(A1 <- A + diag(rep(0.1, n)))## [1] 134.8415kappa(A2 <- A + diag(rep(1, n)))## [1] 15.01074Implementation
sol1 = cg(A1, b, eps = 1e-12)sol1$niter## [1] 78max(abs(A1 %*% sol1$x - b))## [1] 1.588243e-13sol2 = cg(A2, b, eps = 1e-12)sol2$niter## [1] 29max(abs(A2 %*% sol2$x - b))## [1] 1.858513e-13Implementation
- Plot the residual norms (in log-scale) against iterations
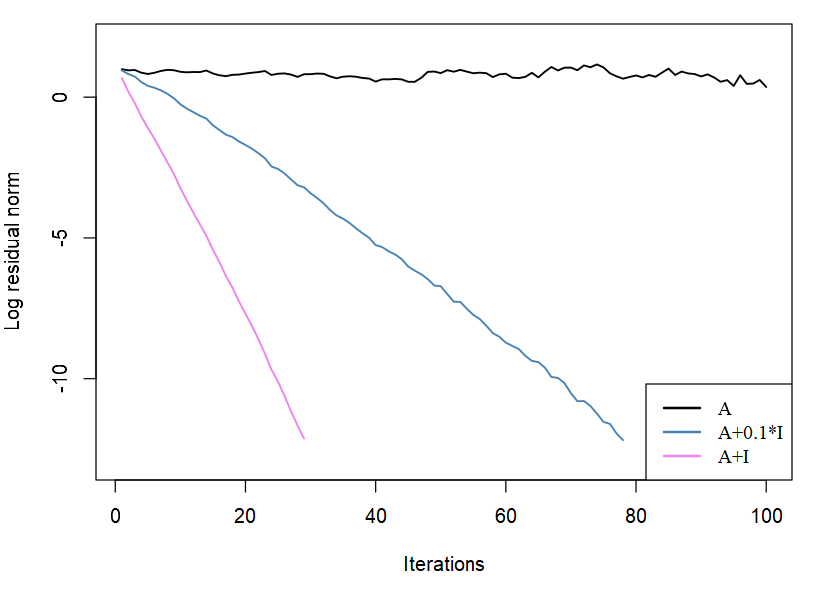
Implementation
- We will see a more useful example when we consider linear regression on sparse matrices
When to Use
A is p.d. and is well-conditioned
The matrix-vector operation v\rightarrow Av is cheap
One example is sparse matrix, or product of sparse matrices
We will elaborate this point when we study sparse matrices
Eigenvalue Computation
Eigenvalues
Eigenvalues are important characteristics of matrices
Wide applications in statistics, e.g., principal component analysis (PCA), spectral clustering, matrix norm, random matrix theory, etc.
Eigenvalue computation is one of the core topics of numerical linear algebra
Definition
The definition of eigenvalue is actually quite simple
For a square matrix A, we call the solution \lambda to the equation Ax=\lambda x an eigenvalue of A
The corresponding x vector is an eigenvector
Here we focus on real symmetric matrices
Decomposition Theorem [2]
A matrix A_{n\times n} is real symmetric if and only if there exists a real orthogonal matrix \Gamma and real eigenvalues \lambda_1,\ldots,\lambda_n such that A=\Gamma D\Gamma', where D=\mathrm{diag}(\lambda_1,\ldots,\lambda_n).
This is typically called the eigenvalue decomposition, or spectral decomposition, of A.
Basic Properties
Suppose that A is a real symmetric matrix
A is p.d. if all its eigenvalues are strictly positive
A^k=\Gamma D^k\Gamma', k=\ldots,-2,-1,0,1,2,\ldots
\det(A)=\lambda_1\cdots\lambda_n
\mathrm{tr}(A)=\lambda_1+\cdots+\lambda_n
Basic Properties
Min-Max principle \lambda_{\min}(A)=\underset{x\neq 0}{\min}\frac{x'Ax}{x'x}=\underset{\Vert x\Vert=1}{\min}\ x'Ax \lambda_{\max}(A)=\underset{x\neq 0}{\max}\frac{x'Ax}{x'x}=\underset{\Vert x\Vert=1}{\max}\ x'Ax
This gives PCA a nice interpretation: the first principal component maximizes the explained variance
Computing Eigenvalues
There are different cases of computing eigenvalues
Computing all eigenvalues (eigenvalue decomposition)
Computing the largest eigenvalue
Computing the eigenvalue that is closest to a given number
...
Computing Eigenvalues
Eigenvalue computation is a large field in numerical linear algebra
The actual algorithms used in practice are usually quite involved
For the purpose of statistical computing, we mainly introduce the basic ideas behind these algorithms and some recommended implementations
In what follows, we focus on finding eigenvalues of real symmetric matrices
All Eigenvalues
Computing All Eigenvalues
The most commonly-used method to compute all eigenvalues consists of two steps:
- Looking for an orthogonal matrix Q such that Q'AQ is tridiagonal
Q'AQ=T=\begin{bmatrix}\alpha_{1} & \beta_{2}\\ \beta_{2} & \alpha_{2} & \beta_{3}\\ & \ddots & \ddots & \ddots\\ & & \beta_{n-1} & \alpha_{n-1} & \beta_{n}\\ & & & \beta_{n} & \alpha_{n} \end{bmatrix}
- Applying the QR algorithm (introduced later) to the matrix T to compute all eigenvalues
The Householder (Tridiagonalization) Algorithm
The Householder algorithm finds a sequence of (simple) orthogonal matrices Q_k such that T=(Q_1\cdots Q_{n-2})'A(Q_1\cdots Q_{n-2}), where T is tridiagonal
Q=Q_1\cdots Q_{n-2} is also orthogonal (why?)
A and T have the same eigenvalues (why?)
The motivation is that T has cheaper matrix operations
The Householder (Tridiagonalization) Algorithm
- A very nice intro orthogonal transformatioduction to the Householder algorithm can be found at Section 4.3 and Section 4.6.1 of https://people.inf.ethz.ch/arbenz/ewp/Lnotes/chapter4.pdf
The QR Algorithm
The QR algorithm iteratively reduces the tridiagonal T into a diagonal matrix by orthogonal transformations:
Initialize T^{(0)}=T, and begin the loop k=0,1,\ldots
Compute the QR decomposition of the matrix T^{(k)}: T^{(k)}=Q_k R_k
Update T^{(k+1)}=R_k Q_k=Q_k'T^{(k)}Q_k
Assume that |\lambda_1|>\cdots>|\lambda_n|>0, and then T^{(k)} converges to a diagonal matrix, whose diagonal elements are eigenvalues of T and A.
Practical Algorithm
- The lower triangular elements T_{ij}^{(k)}, i>j converge to zero at the rate of O(|\lambda_i/\lambda_j|) [2]
- To accelerate the convergence, a shift can be applied to the original matrix:
- Compute the QR decomposition of the shifted matrix, T^{(k)}-\sigma_k I_n=Q_k R_k
- Update T^{(k+1)}=R_k Q_k+\sigma_k I_n=Q_k'T^{(k)}Q_k
- We can take \sigma_k=(T^{(k)})_{nn} until T_{n-1,n}^{(k)}\approx 0, and then switch to \sigma_k=(T^{(k)})_{n-1,n-1}
Complexity
The Householder algorithm costs O(n^3)
The QR decomposition of tridiagonal matrices has a specialized algorithm, which costs O(n^2) instead of O(n^3) (the latter for general dense matrices)
It can be proved that all T^{(k)} matrices are tridiagonal
The number of iterations required in the QR algorithm depends on the distribution of eigenvalues
Largest Eigenvalue
Computing the Largest Eigenvalue
In many applications, we only need the largest eigenvalue (in absolute value)
It can be extended to computing the largest k eigenvalues
If \lambda_1,\ldots,\lambda_n are eigenvalues of A with |\lambda_1|>\cdots>|\lambda_n|, and \gamma_1,\ldots,\gamma_n are the associated eigenvectors, then:
- \lambda_2 is the largest eigenvalue of A-\lambda_1\gamma_1\gamma_1'
- \lambda_3 is the largest eigenvalue of A-\lambda_1\gamma_1\gamma_1'-\lambda_2\gamma_2\gamma_2'
- ...
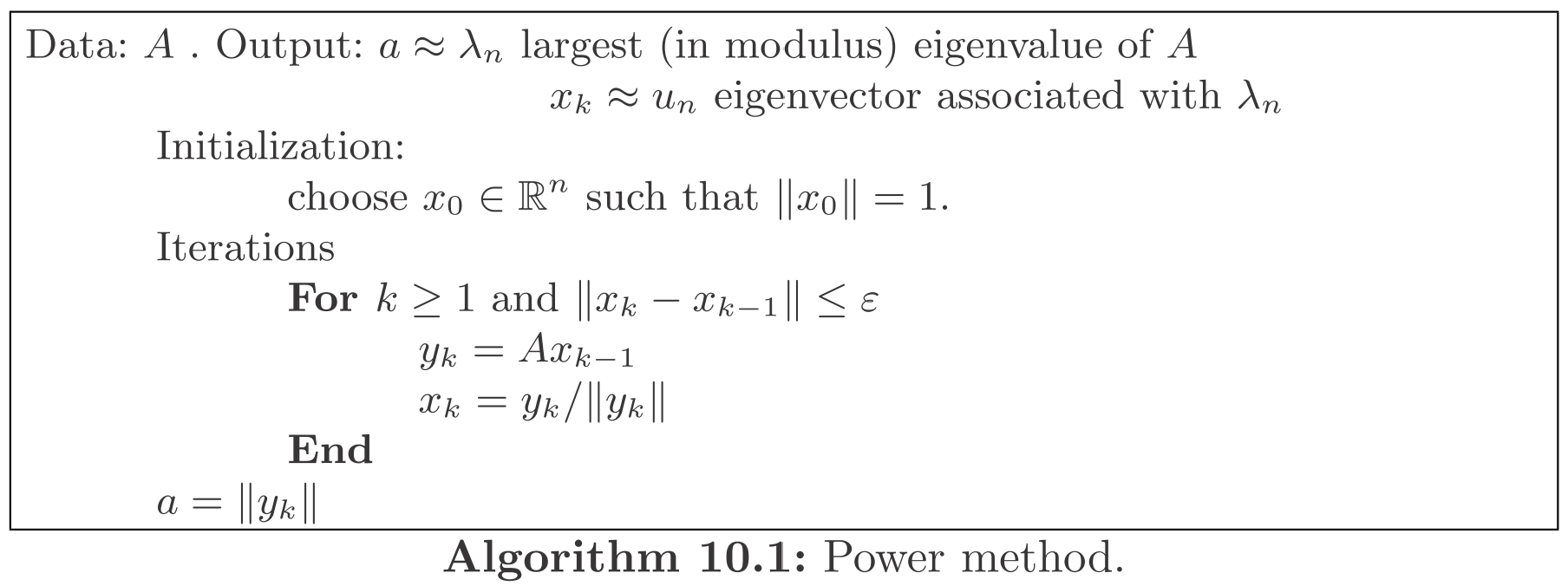
Power Method [2]
One widely-used and possibly the simplest method to compute the largest eigenvalue (in absolute value) is the power method
Convergence [2]
Assume A has eigenvalues \lambda_1>|\lambda_2|\ge\cdots\ge|\lambda_n| and associated eigenvectors \gamma_1,\ldots,\gamma_n. Also assume the initial vector satisfies x_0=\sum_{i=1}^n\beta_i\gamma_i for some \beta_1\neq 0. Then
\left|\Vert y_{k}\Vert-\lambda_{1}\right|\le C\left(\frac{|\lambda_{2}|}{\lambda_{1}}\right)^{k},\qquad\Vert x_{k}-\tilde{\gamma}_{1}\Vert\le C\left(\frac{|\lambda_{2}|}{\lambda_{1}}\right)^{k}, where \tilde{\gamma_1}=\pm\gamma_1.
Pros and Cons
The power method only requires the matrix-vector multiplication v\rightarrow Av to compute eigenvalues of A (recall the CG method)
This is important and useful for sparse matrices
However, it does not effectively use the information of past iterations
Also, it can only compute one eigenvalue at one time
Advanced Algorithms
Mainstream software packages typically use other advanced algorithms to compute the largest eigenvalues, e.g.,
- Implicitly restarted Lanczos method [4]
- Jacobi–Davidson algorithm [5]
They also only need the v\rightarrow Av operation
Can compute multiple eigenvalues together
These algorithms themselves are quite sophisticated, however
Eigenvalue Closest to \mu
Computing Eigenvalue Closest to \mu
Another type of problem is to find the eigenvalue that is closest to a given number \mu
For example, if \mu=0, then this is equivalent to finding the smallest eigenvalue (in absolute value)
Fortunately, we can make use of existing methods that compute largest eigenvalues for this task, via a technique called spectral transformation
Spectral Transformation
Spectral transformation is based on the following finding.
If \lambda_i, i=1,\ldots,n are eigenvalues of A, then Ax_i=\lambda_i x_i for the eigenvectors x_i. Choose a shift \mu\neq\lambda_i, then the eigenvalues of B=(A-\mu I_n)^{-1} are \theta_i=(\lambda_i-\mu)^{-1}, and B x_i=\theta_i x_i.
Therefore, the largest eigenvalue of B corresponds to the eigenvalue of A that is closest to \mu.
Inverse Iteration
If we want to compute the eigenvalue of A closest to \mu
We can apply the power method (or other methods based on matrix-vector multiplication) to B=(A-\mu I_n)^{-1}
The iteration becomes \begin{align*} y_{k} & =(A-\mu I_{n})^{-1}x_{k-1}\\ x_{k} & =y_{k}/\Vert y_{k}\Vert \end{align*}
By the convergence theorem, we have \Vert y_k \Vert\rightarrow \theta_1
Then recover the eigenvalue of A by \lambda_\mu=\mu+1/\theta_1
Solving Linear Systems
In actual implementation, we do not directly compute the matrix inverse (A-\mu I_{n})^{-1}
Instead, we factorize A-\mu I_{n} once using LU decomposition or LDL^T decomposition
And then solve the linear systems (A-\mu I_n)y_k=x_{k-1}
Implementation
The
eigen()function in base R is a very stable and efficient function to compute all eigenvaluesSpecify
symmetric = TRUEif you know your matrix is symmetric (by default it will detect symmetry)Specify
only.values = TRUEif you do not need eigenvectors
Benchmark
library(dplyr)set.seed(123)n = 1000M = matrix(rnorm(n^2), n)A = M + t(M)bench::mark( eigen(A), eigen(A, symmetric = FALSE), eigen(A, symmetric = TRUE), eigen(A, symmetric = TRUE, only.values = TRUE), min_iterations = 3, max_iterations = 10, check = FALSE) %>% select(expression, min, median)## # A tibble: 4 × 3## expression min median## <bch:expr> <bch:tm> <bch:tm>## 1 eigen(A) 1.22s 1.22s## 2 eigen(A, symmetric = FALSE) 4.32s 4.34s## 3 eigen(A, symmetric = TRUE) 1.18s 1.19s## 4 eigen(A, symmetric = TRUE, only.values = TRUE) 326.67ms 330.35msImplementation
- To compute a subset of eigenvalues up to some selection rule, we have several options
- The RSpectra package implements the implicitly restarted Lanczos method
- The PRIMME package uses the Jacobi-Davidson algorithm
- The irlba package is designed for singular value decomposition (next class), but can also be used to compute the largest eigenvalues
- My personal favorite is the RSpectra package (this is a highly biased choice, of course)
Implementation
- The code below computes three eigenvalues with largest absolute values
library(RSpectra)e = eigs_sym(A, k = 3, which = "LM")e$values## [1] 88.83776 88.31500 -88.98945head(e$vectors)## [,1] [,2] [,3]## [1,] 0.01506291 0.029602478 -0.015855700## [2,] -0.05124211 0.016404205 -0.014749483## [3,] -0.01542738 -0.002350839 0.005520141## [4,] 0.02921002 -0.006653817 -0.003387408## [5,] -0.02406133 0.039802584 0.028103112## [6,] -0.01309102 -0.026105929 0.073813795Implementation
- If you want the largest three eigenvalues without taking absolute values, use
which = "LA"
e = eigs_sym(A, k = 3, which = "LA")e$values## [1] 88.83776 88.31500 87.18646- If no eigenvectors are needed, add the following option
e = eigs_sym(A, k = 3, which = "LA", opts = list(retvec = FALSE))e$vectors## NULLBenchmark
- If only part of the eigenvalues are needed,
eigs_sym()is much faster thaneigen()
bench::mark( eigen(A, symmetric = TRUE), eigen(A, symmetric = TRUE, only.values = TRUE), eigs_sym(A, k = 3, which = "LM"), eigs_sym(A, k = 3, which = "LM", opts = list(retvec = FALSE)), min_iterations = 3, max_iterations = 10, check = FALSE) %>% select(min, median)## # A tibble: 4 × 2## min median## <bch:tm> <bch:tm>## 1 1.2s 1.2s## 2 330.7ms 335.7ms## 3 158.8ms 159.2ms## 4 157.1ms 159.3ms- The difference will be more evident on sparse matrices (next class)
Implementation
- If one needs the smallest three eigenvalues (in absolute value)
e = eigs_sym(A, k = 3, which = "LM", sigma = 0)e$values## [1] 0.01589015 -0.14248784 -0.26307129In general, if
sigmais notNULL, then the selection rule is applied to 1/(\lambda_i-\sigma)This only affects the selection rule
The returned eigenvalues are still for A
References
[1] Folkmar Bornemann (2018). Numerical Linear Algebra. Springer.
[2] Grégoire Allaire and Sidi Mahmoud Kaber (2008). Numerical linear algebra. Springer.
[3] Åke Björck (2015). Numerical methods in matrix computations. Springer.
[4] Danny C. Sorensen (1997). Implicitly restarted Arnoldi/Lanczos methods for large scale eigenvalue calculations. In Parallel Numerical Algorithms (pp. 119-165). Springer.
[5] Gerard L.G. Sleijpen and Henk A. Van der Vorst (1996). A Jacobi–Davidson iteration method for linear eigenvalue problems. SIAM Journal on Matrix Analysis and Applications, 17(2), 401-425.