Computational Statistics
Lecture 3
Yixuan Qiu
2023-09-20
Numerical Linear Algebra
Today's Topics
Case studies
Spline fitting
Back to regression
New topics
LDLT decomposition
QR decomposition
Spline Fitting
Interpolation Problem
Given observed points x=(x0,…,xn)′ and y=(y0,…,yn)′, find a function f(x) such that f(xi)=yi,i=0,…,n.
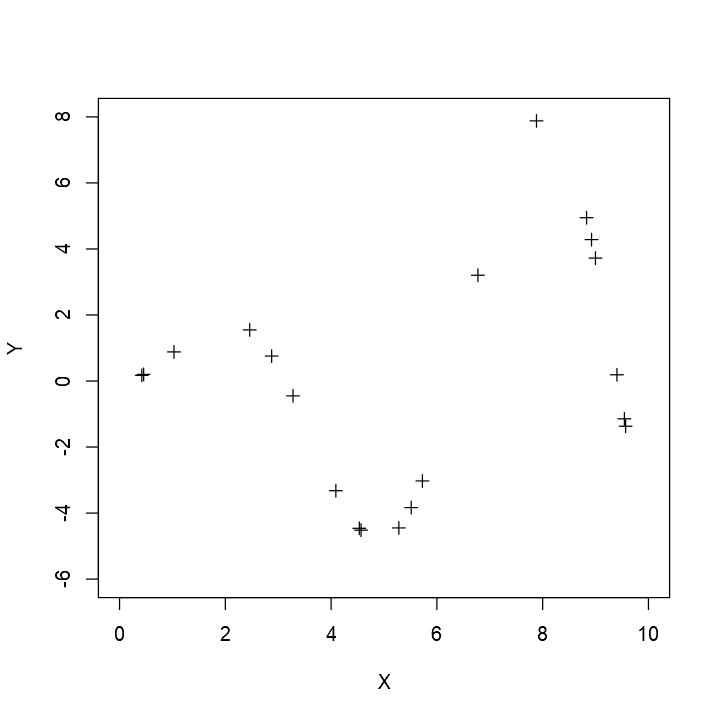
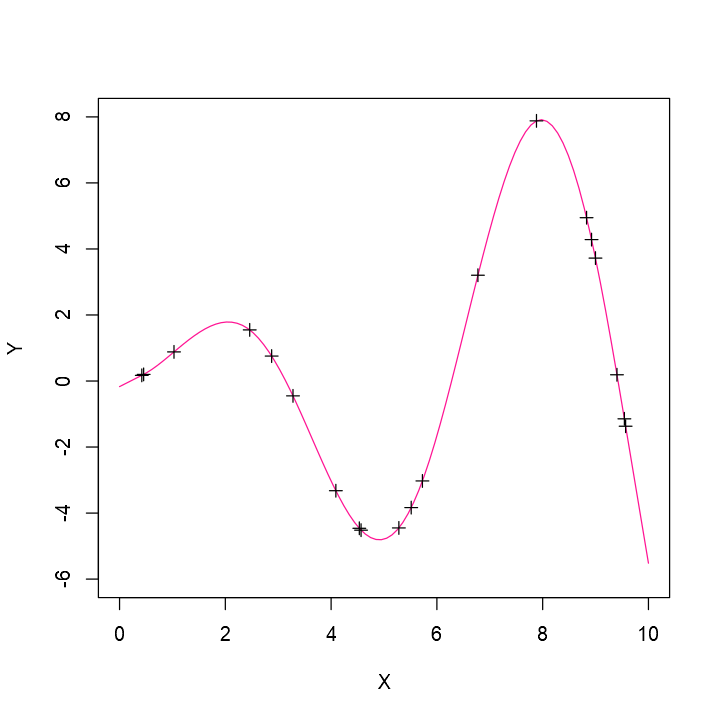
Spline Fitting
Without loss of generality we can assume that x0<x1<⋯<xn
We typically require that f is continuous, or differentiable, or smooth (2nd derivative exists)
One important class of functions commonly used for interpolation are piecewise polynomials, also known as spline functions
Spline fitting is the task of determining polynomial coefficients in each interval
Natural Cubic Spline
A commonly-used spline is the natural cubic spline
Has continuous second derivative
On each interval [xi,xi+1], it is a cubic polynomial f(x)=fi(x)=ai+bi(x−xi)+ci(x−xi)2+di(x−xi)3
We need to determine the coefficients (ai,bi,ci,di), i=0,…,n−1
Natural Cubic Spline
- Natural cubic splines must satisfy some constraints
- Interpolating observed points
- f0(x0)=y0
- fi−1(xi)=fi(xi)=yi, i=1,…,n−1
- fn−1(xn)=yn
- Continuous first derivative
- f′i−1(xi)=f′i(xi), i=1,…,n−1
- Continuous second derivative
- f′′i−1(xi)=f′′i(xi), i=1,…,n−1
- Zero second derivative at end points
- f′′0(x0)=f′′n−1(xn)=0
Theorem
The natural cubic spline that satisfies the conditions in the previous slide has the form
fi(x)=zi+1(x−xi)3+zi(xi+1−x)36hi+(yi+1hi−zi+1hi6)(x−xi)+(yihi−zihi6)(xi+1−x),i=0,…,n−1 where hi=xi+1−xi, and zi=f′′(xi) are values to be determined
Spline Fitting
We have assume that z0=zn=0
So there are (n−1) unknown values z1,…,zn−1
The relation f′i−1(xi)=f′i(xi), i=1,…,n−1 gives 6(bi−bi−1)=hi−1zi−1+2(hi−1+hi)zi+hizi+1, where bi=(yi+1−yi)/hi
Spline Fitting
Further let vi=2(hi−1+hi), ui=6(bi−bi−1)
Then we get a tridiagonal system
⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣v1h1h1v2h2h2v3⋱⋱⋱hn−2hn−2vn−1⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣z1z2z3⋮zn−1⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦=⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣u1u2u3⋮un−1⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦
- Solving the system gives the expression of the spline function
Back to Regression
Computing predicted values of linear regression ^y=X(X′X)−1X′Y.
NEVER do the following:
X %*% solve(t(X) %*% X) %*% t(X) %*% Y- Benchmark
set.seed(123)X = matrix(rnorm(5000 * 500), 5000)Y = rnorm(5000)system.time(X %*% solve(t(X) %*% X) %*% t(X) %*% Y)## user system elapsed ## 9.09 0.03 9.36- What's wrong with this implementation?
First improvement
- Let's first try a simple "magic"
system.time(X %*% (solve(t(X) %*% X) %*% (t(X) %*% Y)))## user system elapsed ## 0.91 0.00 0.93- Recall the previous implementation
X %*% solve(t(X) %*% X) %*% t(X) %*% YYou get ~10x speedup almost for free
The only differences are two pairs of parentheses!
Principle 1
When multiplying several matrices together, prefer to first compute the product involving smaller matrices
Reason: the computational complexity of the matrix multiplication Am×nBn×p is O(mnp)
Second improvement
system.time(X %*% solve(t(X) %*% X, t(X) %*% Y))## user system elapsed ## 0.78 0.00 0.76Principle 2
Avoid explicit matrix inversion whenever possible
Instead, solve linear systems
In fact, in statistical computing, it is very rare that you really need A−1
Computing A−1B is equivalent to solving AX=B
Computing BA−1 is equivalent to solving A′X=B′
Third improvement
system.time(X %*% solve(crossprod(X), crossprod(X, Y)))## user system elapsed ## 0.72 0.00 0.70Principle 3
Avoid explicit matrix transpose
Try specialized functions
crossprod(x): compute X′Xcrossprod(x, y): compute X′Ytcrossprod(x, y): compute XY′
Principle 4
Utilize special structures of matrices
For example, symmetry, positive definiteness, etc.
Try to further improve the solution!
LDLT Decomposition
Motivation
LU decomposition applies to general square matrices
Cholesky decomposition applies to positive definite matrices
- Halves the storage cost
- Typically faster than LU decomposition
If we only know the matrix is symmetric
Is there any tailored decomposition method?
LDLT Decomposition
[4] shows that a nonsingular symmetric matrix A can be decomposed as A=LDL′
L is lower triangular with ones on the diagonal
D is a block diagonal matrix, each block being 1×1 or 2×2, e.g.,
⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣1−34−40−310−126134−1211−1−25−46−16−5013−25542⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦=⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣1−314−21−40−3103521⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣1122−1−12⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦⎡⎢ ⎢ ⎢ ⎢ ⎢ ⎢⎣1−34−401−2031−35121⎤⎥ ⎥ ⎥ ⎥ ⎥ ⎥⎦
- In practice, a permutation matrix would be applied to A for factorization, i.e., P′AP=LDL′
Solving Linear Systems
Suppose the decomposition P′AP=LDL′ is given
If we wish to solve Ax=b, then:
Compute ~b=P′b
Solve Ly1=~b to get y1
Solve Dy2=y1 to get y2
Solve L′~x=y2 to get ~x
Compute x=P~x
Solving Linear Systems
Solving Dy=c is similar to solving a diagonal system
Since D is block diagonal, D=bdiag(D1,…,DK)
Each Dk is 1×1 or 2×2
Partition c according to D, c=(c′1,…,c′K)′
Then y=((D−11c1)′,…,(D−1KcK)′)′
If Dk is 2×2, use closed-form inverse formula
Algorithm
The most commonly-used algorithm for the LDLT decomposition is the Bunch-Kaufman algorithm [5]
We omit the details here since it is pretty involved
Interested readers are referred to [4] and [5]
The computational complexity is O(n3), with a multiplicative factor similar to Cholesky decomposition
Implementation
Unfortunately, at the current moment there is no mature R interface to this decomposition
There is indeed a
BunchKaufman()function in the Matrix package, but it is somewhat incompleteThe good news is that the algorithm has been implemented in the LAPACK library, which is included by R
We just need to write a simple C++ wrapper to call the function via Rcpp
Implementation
- Simple wrapper for the factorization function
ldlt_fac = function(A){ res = Matrix::BunchKaufman(A, uplo = "L") list(perm = res@perm, fac = matrix(res@x, res@Dim[1]))}Implementation
- C++ code to call the solving function
- This will create an R function
ldlt_solve()
compiler_flags = "-lRlapack -lRblas -lgfortran -lm -lquadmath"Sys.setenv("PKG_LIBS" = compiler_flags)Rcpp::cppFunction(includes = "#include <R_ext/Lapack.h>",code = " NumericVector ldlt_solve(List fac, NumericVector b) { NumericMatrix fac_res = fac[\"fac\"]; IntegerVector perm = fac[\"perm\"]; NumericVector sol = Rcpp::clone(b); const int n = fac_res.nrow(); const int nrhs = 1; char uplo = 'L'; int info; F77_CALL(dsytrs)( &uplo, &n, &nrhs, fac_res.begin(), &n, perm.begin(), sol.begin(), &n, &info FCONE); return sol; }")Implementation
- Verify the result
set.seed(123)n = 2000M = matrix(rnorm(n^2), n)A = M + t(M)b = rnorm(n)fac = ldlt_fac(A)x = ldlt_solve(fac, b)max(abs(A %*% x - b)) # Verify A*x = b## [1] 8.482548e-12Benchmark
- This implementation is much faster than
solve()
library(dplyr)bench::mark( solve(A, b), ldlt_fac(A), ldlt_solve(fac, b), max_iterations = 10, check = FALSE) %>% select(expression, min, median)## # A tibble: 3 × 3## expression min median## <bch:expr> <bch:tm> <bch:tm>## 1 solve(A, b) 1.44s 1.44s## 2 ldlt_fac(A) 813.09ms 813.09ms## 3 ldlt_solve(fac, b) 3.44ms 4.09msWhen to Use?
As long as you can guarantee A is symmetric, use LDLT decomposition to solve linear systems
If positive definiteness is guaranteed, use Cholesky decomposition
QR Decomposition
Motivation
Most of the decomposition methods introduced before heavily rely on triangular matrices
This is because triangular matrices have cheap "inverse" operations
Another special type of matrices is the orthogonal matrix
Q′Q=QQ′=I, which implies Q−1=Q′
QR Decomposition
QR decomposition factorizes a square matrix into the product of two matrices, A=QR
Q is an orthogonal matrix, Q′Q=QQ′=I
R is upper triangular
Factorization Theorem [2]
Let A be a nonsingular square matrix. There exists a unique pair (Q,R), where Q is an orthogonal matrix and R is an upper triangular matrix with positive diagonal entries, such that A=QR.
Solving Linear Systems
Suppose the decomposition A=QR is given
If we wish to solve Ax=b, then:
First, compute ~b=Q′b
Second, solve Rx=~b to get x
Solving Linear Systems
Suppose the decomposition A=QR is given
If we wish to solve Ax=b, then:
First, compute ~b=Q′b
Second, solve Rx=~b to get x
However, it is uncommon to directly use QR decomposition for solving linear systems
Solving Linear Systems
Suppose the decomposition A=QR is given
If we wish to solve Ax=b, then:
First, compute ~b=Q′b
Second, solve Rx=~b to get x
However, it is uncommon to directly use QR decomposition for solving linear systems
Typically slower than LU decomposition
More useful in factorizing rectangular matrices
Factorization Theorem [3]
Let Am×n be a matrix with rank(A)=n, n≤m. Then there exist an orthogonal matrix Qm×m and an upper triangular matrix Rn×n with positive diagonal entries, such that A=Q(R0)=(Q1Q2)(R0). In addition, the matrices R and Q1=AR−1 are uniquely determined.
Thin QR Decomposition
According to the factorization theorem in the previous slide, any rectangular matrix A with full column rank can be factorized as A=Q1R
A∈Rm×n, Q1∈Rm×n, R∈Rn×n
Q1 is column-orthonormal, meaning that Q′1Q1=In×n
This is called the thin QR decomposition
Relation with Cholesky Decomposition
If the (thin) QR decomposition result X=QR is given
Then X′X=R′Q′QR=R′R, which is exactly the Cholesky decomposition of A=X′X
Relation with Regression
To see why the thin QR decomposition is useful, recall the regression coefficient estimate ^β=(X′X)−1X′Y and predicted values ^y=X^β
Suppose we have Xn×p=QR, X full column rank, then:
- ^β=(R′R)−1R′Q′Y=R−1Q′Y
- ^y=QR^β=QQ′Y
QQ′ is essentially the projection matrix, or hat matrix, in regression analysis
Relation with Regression
To see why the thin QR decomposition is useful, recall the regression coefficient estimate ^β=(X′X)−1X′Y and predicted values ^y=X^β
Suppose we have Xn×p=QR, X full column rank, then:
- ^β=(R′R)−1R′Q′Y=R−1Q′Y
- ^y=QR^β=QQ′Y
QQ′ is essentially the projection matrix, or hat matrix, in regression analysis
Warning: Qn×p is only column-orthonomal, so QQ′≠In×n! (We do have Q′Q=Ip×p)
Algorithm
Here we omit the computing algorithm for QR decomposition
Interested readers are referred to
- Section 9 of [1] (modified Gram-Schmidt algorithm)
- Section 7.3.4 of [2] (Householder algorithm)
- Section 2.3.2 of [3] (Householder algorithm)
The important point is, in practice we do NOT explicitly form the Q matrix
Instead, given a vector y, computing Qy and Q′y has specialized algorithms
Computational Complexity
For rectangular matrix Xn×p, X full column rank, n≥p
Computing the thin QR decomposition using the modified Gram-Schmidt algorithm costs O(np2)
Using the Householder algorithm costs O(np2−p3/3)
Implementation
- The
qr()function in R can perform QR decomposition
set.seed(123)x = matrix(rnorm(25), 5, 5)qr_res = qr(x)Implementation
- The result is a compact representation of Q and R
qr_res## $qr## [,1] [,2] [,3] [,4] [,5]## [1,] 1.67880106 -1.8735119 -0.1240544 -2.4977185 -0.6449736## [2,] 0.13710826 -1.3836930 -1.2267898 -0.6009298 0.7682812## [3,] -0.92846517 0.8909949 -0.7676374 -1.1355032 0.8695156## [4,] -0.04199925 -0.4147299 0.9330131 0.3672150 -1.0253122## [5,] -0.07701195 -0.1723435 -0.3178735 -0.8619799 -0.5906208## ## $rank## [1] 5## ## $qraux## [1] 1.3338547 1.0665197 1.1686505 1.5069425 0.5906208## ## $pivot## [1] 1 2 3 4 5## ## attr(,"class")## [1] "qr"Implementation
- To get the explicit Q and R matrices, use
qr.Q()andqr.R()
qr.Q(qr_res)## [,1] [,2] [,3] [,4] [,5]## [1,] -0.33385471 -0.7874465 -0.2822091 0.43404722 -0.02073790## [2,] -0.13710826 -0.1474621 -0.2109068 -0.47031583 0.83293313## [3,] 0.92846517 -0.3428718 -0.1241733 0.01466849 0.06897252## [4,] 0.04199925 0.4395243 -0.8533937 0.27599706 0.02448081## [5,] 0.07701195 0.2178078 0.3635610 0.71694943 0.54812026qr.R(qr_res)## [,1] [,2] [,3] [,4] [,5]## [1,] 1.678801 -1.873512 -0.1240544 -2.4977185 -0.6449736## [2,] 0.000000 -1.383693 -1.2267898 -0.6009298 0.7682812## [3,] 0.000000 0.000000 -0.7676374 -1.1355032 0.8695156## [4,] 0.000000 0.000000 0.0000000 0.3672150 -1.0253122## [5,] 0.000000 0.000000 0.0000000 0.0000000 -0.5906208Implementation
- But you rarely need to explicitly form Q
- To compute Qv and Q′v, use
qr.qy()andqr.qty()
Q = qr.Q(qr_res)v = rnorm(5)Q %*% v## [,1]## [1,] -0.65989192## [2,] 1.65499813## [3,] -1.80255035## [4,] -0.11692776## [5,] -0.02040374qr.qy(qr_res, v)## [1] -0.65989192 1.65499813 -1.80255035 -0.11692776 -0.02040374Implementation
crossprod(Q, v) # == t(Q) %*% v## [,1]## [1,] 0.6394024## [2,] 0.9249034## [3,] 1.7073775## [4,] -0.5390799## [5,] 1.4027564qr.qty(qr_res, v)## [1] 0.6394024 0.9249034 1.7073775 -0.5390799 1.4027564Implementation
- The
qr()function also supports thin QR decomposition
set.seed(123)x = matrix(rnorm(15), 5, 3)qr_res = qr(x)qr.Q(qr_res)## [,1] [,2] [,3]## [1,] -0.33385471 -0.7874465 -0.2822091## [2,] -0.13710826 -0.1474621 -0.2109068## [3,] 0.92846517 -0.3428718 -0.1241733## [4,] 0.04199925 0.4395243 -0.8533937## [5,] 0.07701195 0.2178078 0.3635610qr.R(qr_res)## [,1] [,2] [,3]## [1,] 1.678801 -1.873512 -0.1240544## [2,] 0.000000 -1.383693 -1.2267898## [3,] 0.000000 0.000000 -0.7676374Implementation
- You can also get the full QR decomposition by adding the
complete = TRUEargument
qr.Q(qr_res, complete = TRUE)## [,1] [,2] [,3] [,4] [,5]## [1,] -0.33385471 -0.7874465 -0.2822091 -0.23791265 0.36362704## [2,] -0.13710826 -0.1474621 -0.2109068 0.95639468 0.01684644## [3,] 0.92846517 -0.3428718 -0.1241733 0.05201684 0.04760905## [4,] 0.04199925 0.4395243 -0.8533937 -0.11881268 0.25031427## [5,] 0.07701195 0.2178078 0.3635610 0.10901648 0.89586144qr.R(qr_res, complete = TRUE)## [,1] [,2] [,3]## [1,] 1.678801 -1.873512 -0.1240544## [2,] 0.000000 -1.383693 -1.2267898## [3,] 0.000000 0.000000 -0.7676374## [4,] 0.000000 0.000000 0.0000000## [5,] 0.000000 0.000000 0.0000000Benchmark
library(Matrix)set.seed(123)M = matrix(rnorm(2000^2), 2000)A = crossprod(M)bench::mark( lu(A), chol(A), qr(A), max_iterations = 10, check = FALSE) %>% select(expression, min, median)## # A tibble: 3 × 3## expression min median## <bch:expr> <bch:tm> <bch:tm>## 1 lu(A) 1.32s 1.32s## 2 chol(A) 1.27s 1.27s## 3 qr(A) 4.18s 4.18sBenchmark
set.seed(123)n = 2000p = 200x = matrix(rnorm(n * p), n, p)bench::mark( chol(crossprod(x)), qr(x), max_iterations = 100, check = FALSE) %>% select(expression, min, median)## # A tibble: 2 × 3## expression min median## <bch:expr> <bch:tm> <bch:tm>## 1 chol(crossprod(x)) 42.2ms 45.1ms## 2 qr(x) 57.6ms 58.2msWhen to Use?
QR decomposition is seldom used to solve linear systems directly
- It is typically slower than LU decomposition
Thin QR decomposition is widely used in regression computation
- It is slightly slower than Cholesky decomposition on X′X
- But QR decomposition is more stable
References
[1] Folkmar Bornemann (2018). Numerical Linear Algebra. Springer.
[2] Grégoire Allaire and Sidi Mahmoud Kaber (2008). Numerical linear algebra. Springer.
[3] Åke Björck (2015). Numerical methods in matrix computations. Springer.
[4] James R. Bunch and Beresford N. Parlett (1971). Direct methods for solving symmetric indefinite systems of linear equations. SIAM Journal on Numerical Analysis.
[5] James R. Bunch and Linda Kaufman (1977). Some stable methods for calculating inertia and solving symmetric linear systems. Mathematics of computation.